This article describes the development of an artificial intelligence model that can identify about 460 Finnish true bug species (luteet; Heteroptera) from photographs. The resulting web application, available at taxai.biomi.org, provides a tool to assist with species identification by suggesting the most likely species based on an image.
The project aims to complement—rather than replace—human expertise in species identification. While the model achieves 86% accuracy based on testing, it should be viewed as a helpful first step in the identification process rather than a definitive authority.
The following sections detail the technical process of creating this identification tool, from data collection through model training to public deployment.
Collecting Images for Training
I began by getting a list of Finnish true bug species from Laji.fi (560 species). I then made a set of Python scripts to systematically download species photographs (jpg, jpeg, or png) from multiple sources via their APIs:
- Laji.fi observations: I excluded entries with quality issues, specimens in egg lifestage, iNaturalist observations (handled separately), and Löydös open form observations (which have lower reliability compared to other sources)
- iNaturalist taxon photos: These typically represent high-quality, curated images for each species
- iNaturalist observations: Limited to research-grade observations within 5000 kilometers of Helsinki without conflicting identifications.
Downloading the images took several days due to throttling to avoid overloading the services. From iNaturalist, I retrieved large-format images (second-highest resolution), while from Laji.fi I downloaded original images. Images were organized hierarchically by source and species, with each file named according to the pattern ”Species_name_identifier”. This ensured unique filenames while keeping species names for easier data handling.
For each image, I generated corresponding JSON metadata files containing details such as source, species name, image ID, author, and lifestage. So far, I haven’t used these metadata files.
My scripts were set to download up to 200 photographs per species, prioritizing sources in the following order: iNaturalist taxon photos (typically 2-5 per species), Laji.fi observations, and finally iNaturalist observation photos. Species with abundant representation in Laji.fi required fewer additional images from iNaturalist observations. Overall, I gathered images of 744 species, exceeding the 560 Finnish species target due to taxonomic naming variations. Some species had more than 200 photos, due to e.g. testing different versions of the scripts fetching the photos.
I reviewed all images by quickly looking for outliers – photos that won’t help with automatic identification. This took a few hours. I copied about 560 problematic files to a separate folder for later filtering. Most outliers were microscope photos of small details or preparations, habitat images, and specimen labels. I likely missed some outliers during this process.
Dataset Preparation
Next, I created a list of image paths and their species labels for model training. During this step, I filtered out:
- The outlier images I found earlier
- Duplicate photos (appearing in both iNaturalist taxon and observation datasets)
- Species not found in Finland
- Images that were too small (less than 200 pixels in either dimension; there weren’t many)
The dataset was very unbalanced: common species had many photos while rare species had only few. About half of the Finnish species had fewer than 80 images, and 10% had fewer than 13 images. For good training results, the ratio between common and rare species should be no more than 10 to 1, even though real-world data is unbalanced. This made image augmentation necessary.
Data Augmentation
I decided to create 15 augmented images per species. This would roughly double the number of images for rare species but increase the count by less than 10% for common ones. For comparison, iNaturalist requires at least 20 images per species before including it in their identification model, so I aimed for a roughly similar number. I limited augmentation so each original image was used only once, with images for augmentation selected randomly.
The augmentation process:
- Used the
rembgPython library with theisnet-general-usemodel to detect the most important part of the photo and remove background (aka salient object detection). I chose this model and settings based on what looked good – it separates most insects from backgrounds without making the edges too sharp. - Rotated each photo randomly after removing the background
- Added a new background from a set of 10 options (plain colors, blurred textures, and blurred vegetation), stretched to fit the image size
- Randomly flipped some images
- Zoomed in 0-20%
- Made subtle random adjustments to brightness, contrast, and saturation
- Added slight random blur and noise
I saved each augmented image with ”_AUGMENTED” in the filename to clearly distinguish between original and transformed images. This process struggled with well-camouflaged insects, like Aradus species on tree bark.
Example of original (left) and augmented image:

Tips for Data Processing Scripts
When creating scripts for major data work:
- Make them resumable – they should be able to stop and restart where they left off. Something will always go wrong with scripts that run for hours or days, so starting over each time wastes time.
- Clearly mark what’s changed and what hasn’t. Make copies of new data, keep the originals intact, and label both clearly.
- Use Git for version control of your scripts and track which script version was used to process which data set.
- Have a “single source of truth” for all data, to avoid data inconsistency when data is copied between computers or tools. (This is why I made copies of the outlier images instead of just removing them; the image files themselves are the source of truth of what has been downloaded, augmented or excluded.)
Model Training and Testing
Next, I created a list of all images for training. I only included species with at least 30 images, giving me about 60,000 images across 462 species (list of species here). I randomly set aside 1% of images for later testing of model accuracy. Of the remaining images, I used 85% for training and 15% for validation.
For my identification model, I chose EfficientNet that has shown good results e.g. classifying butterfly life stages. After several tests, I selected the B5 version, which uses 456 × 456 pixel images. This is a medium-large model – EfficientNet sizes range from 224 to 600 pixels. Larger models are usually more accurate but slower. Like most models, EfficientNet requires square images, so all photos are resized to square format during the training process. During training, I also applied basic augmentation to randomly rotate, resize, and flip images. This creates variety in the training data to prevent the model from simply memorizing the dataset (aka. overfitting), which improves performance on new images.
Training the model took 27 hours on my desktop computer (NVIDIA RTX 4060 Ti graphics card with 16 GB memory). Testing showed the best model achieved 86% accuracy – meaning its top prediction was correct for 86% of test images.
User Interface and Deployment
The final step was creating a user interface and deploying the model online. I developed this locally using Docker and chose Google Cloud Run for hosting. Cloud Run isn’t specifically designed for AI models, but it’s relatively simple and affordable for small applications like this one.
The app is available at https://taxai.biomi.org – feel free to test it, though it’s still a work in progress. Since the model only knows true bugs, it will try to identify any image you upload as a bug species, even if there’s no bug present.
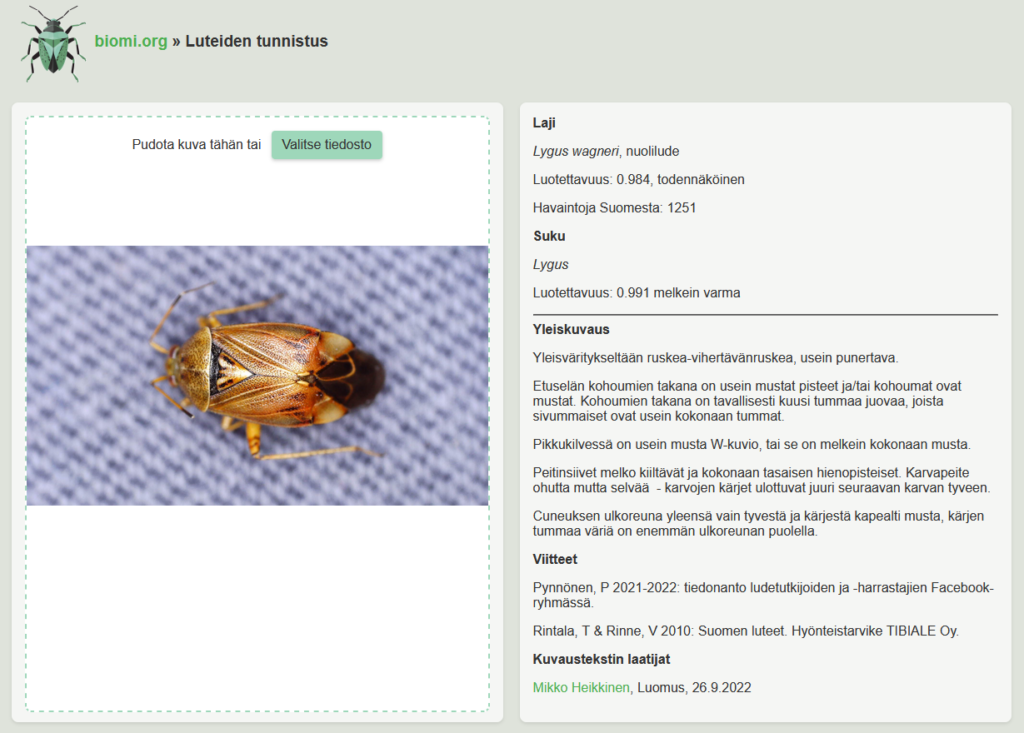
The interface lets you upload an image and shows the prediction results. It also retrieves additional information from the FinBIF API: Finnish name, number of observations (showing if the species is rare or common), and identification notes when available.
Future Improvements
The model could be improved in many ways, e.g.:
- Create another model to detect whether an image contains a bug. This would help filter out irrelevant images from the training data (e.g. habitats), improving accuracy.
- Develop an object detection model to locate bugs within images and zoom in on them. This could significantly improve accuracy when bugs are small in the frame. This could also help identify outlier images.
- Do more extensive image augmentation to increase the training dataset: partial occlusions, simulated overlays (e.g., raindrops), adding noise, zooming with object detection, AugMix, CutMix, or MixUp. There are many different kinds of methods.
- Collect more images especially of rare species to improve performance.
This approach can be adapted to any taxonomic group with sufficient and reliable photographs. While the process isn’t technically difficult, gathering and curating quality training data requires significant time investment. The frontend code is available as open source, with the remaining components to be released in the future. I hope this walkthrough helps others looking to build similar identification tools for their areas of interest.
PS. This project also served as a test case for using modern AI tools throughout development. I wrote most of the code with the Cursor AI code editor, consulted ChatGPT and Claude about model training and configuration options, designed the user interface based on a DALL-E generated concept, created the logo using Midjourney, and had Claude proofread this blog post.